Meta-analysis for milk fat and protein percentage using imputed sequence variant genotypes in 94,321 cattle from eight cattle breeds
Résumé
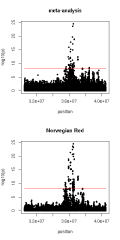
AbstractBackgroundSequence-based genome-wide association studies (GWAS) provide high statistical power to identify candidate causal mutations when a large number of individuals with both sequence variant genotypes and phenotypes is available. A meta-analysis combines summary statistics from multiple GWAS and increases the power to detect trait-associated variants without requiring access to data at the individual level of the GWAS mapping cohorts. Because linkage disequilibrium between adjacent markers is conserved only over short distances across breeds, a multi-breed meta-analysis can improve mapping precision.ResultsTo maximise the power to identify quantitative trait loci (QTL), we combined the results of nine within-population GWAS that used imputed sequence variant genotypes of 94,321 cattle from eight breeds, to perform a large-scale meta-analysis for fat and protein percentage in cattle. The meta-analysis detected (p ≤ 10−8) 138 QTL for fat percentage and 176 QTL for protein percentage. This was more than the number of QTL detected in all within-population GWAS together (124 QTL for fat percentage and 104 QTL for protein percentage). Among all the lead variants, 100 QTL for fat percentage and 114 QTL for protein percentage had the same direction of effect in all within-population GWAS. This indicates either persistence of the linkage phase between the causal variant and the lead variant across breeds or that some of the lead variants might indeed be causal or tightly linked with causal variants. The percentage of intergenic variants was substantially lower for significant variants than for non-significant variants, and significant variants had mostly moderate to high minor allele frequencies. Significant variants were also clustered in genes that are known to be relevant for fat and protein percentages in milk.ConclusionsOur study identified a large number of QTL associated with fat and protein percentage in dairy cattle. We demonstrated that large-scale multi-breed meta-analysis reveals more QTL at the nucleotide resolution than within-population GWAS. Significant variants were more often located in genic regions than non-significant variants and a large part of them was located in potentially regulatory regions.
Domaines
Sciences du Vivant [q-bio]
Fichier principal
 12711_2020_Article_556.pdf (1.77 Mo)
Télécharger le fichier
12711_2020_Article_556.pdf (1.77 Mo)
Télécharger le fichier
 12711_2020_556_MOESM4_ESM.png (6.1 Ko)
Télécharger le fichier
12711_2020_556_MOESM4_ESM.png (6.1 Ko)
Télécharger le fichier
 12711_2020_556_MOESM5_ESM.png (21.51 Ko)
Télécharger le fichier
12711_2020_556_MOESM5_ESM.png (21.51 Ko)
Télécharger le fichier
 12711_2020_556_MOESM6_ESM.png (21.12 Ko)
Télécharger le fichier
12711_2020_556_MOESM1_ESM.docx (285.04 Ko)
Télécharger le fichier
12711_2020_556_MOESM2_ESM.xlsx (42.19 Ko)
Télécharger le fichier
12711_2020_556_MOESM3_ESM.xlsx (56.13 Ko)
Télécharger le fichier
12711_2020_556_MOESM7_ESM.docx (24.11 Ko)
Télécharger le fichier
12711_2020_556_MOESM8_ESM.xlsx (30.86 Ko)
Télécharger le fichier
12711_2020_556_MOESM9_ESM.xlsx (31.53 Ko)
Télécharger le fichier
12711_2020_556_MOESM6_ESM.png (21.12 Ko)
Télécharger le fichier
12711_2020_556_MOESM1_ESM.docx (285.04 Ko)
Télécharger le fichier
12711_2020_556_MOESM2_ESM.xlsx (42.19 Ko)
Télécharger le fichier
12711_2020_556_MOESM3_ESM.xlsx (56.13 Ko)
Télécharger le fichier
12711_2020_556_MOESM7_ESM.docx (24.11 Ko)
Télécharger le fichier
12711_2020_556_MOESM8_ESM.xlsx (30.86 Ko)
Télécharger le fichier
12711_2020_556_MOESM9_ESM.xlsx (31.53 Ko)
Télécharger le fichier
Origine : Publication financée par une institution
Format : Figure, Image
Format : Figure, Image
Format : Figure, Image
Loading...


