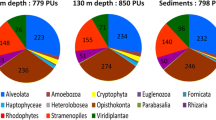
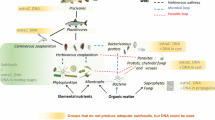
Abstract
Protists are unicellular eukaryotes found in almost all biomes on Earth. Despite their critical importance in aquatic ecosystems, there is still a lack of information about how and if anthropogenic disturbances impact protists diversity and abundance. Improvements in molecular ecology techniques, and more specifically the study of protist communities through sedDNA approaches has greatly improved our understanding of the long-term changes in their diversity, distribution, and relative importance in lake ecosystems. In this chapter, we focus on the application of molecular biological techniques to sedimentary DNA (sedDNA) to investigate a broad temporal perspective on the biodiversity, community assemblages and populations dynamic of lake protists. We demonstrate how their paleo-reconstructions offer a panel of unique information related to their role in lake ecosystems (i.e., remobilization and recycling of carbon and nutrients, grazers and consumers in aquatic food webs), and provide the potential unlock new avenues for paleolimnological investigations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol 66(1):4–119. https://doi.org/10.1111/jeu.12691
Anslan S, Kang W, Dulias K, Wünnemann B, Echeverría-Galindo P, Börner N, Schwarz A, Liu Y, Liu K, Künzel S, Kisand V, Rioual P, Peng P, Wang J, Zhu L, Vences M, Schwalb A (2022) Compatibility of diatom valve records with sedimentary ancient DNA amplicon data: a case study in a brackish, alkaline Tibetan lake. Front Earth Sci 10
Armbrecht L, Herrando-Pérez S, Eisenhofer R, Hallegraeff GM, Bolch CJS, Cooper A (2020) An optimized method for the extraction of ancient eukaryote DNA from marine sediments. Mol Ecol Resour 20(4):906–919. https://doi.org/10.1111/1755-0998.13162
Armbrecht L, Hallegraeff G, Bolch CJS, Woodward C, Cooper A (2021a) Hybridisation capture allows DNA damage analysis of ancient marine eukaryotes. Sci Rep 11:3220. https://doi.org/10.1038/s41598-021-82578-6
Armbrecht L, Eisenhofer R, Utge J, Sibert EC, Rocha F, Ward R, Pierella Karlusich JJ, Tirichine L, Norris R, Summers M, Bowler C (2021b) Paleo-diatom composition from Santa Barbara Basin deep-sea sediments: a comparison of 18S-V9 and diat-rbcL metabarcoding vs shotgun metagenomics. ISME Commun 1(1):1–10. https://doi.org/10.1038/s43705-021-00070-8
Armbrecht L, Weber ME, Raymo ME, Peck VL, Williams T, Warnock J, Kato Y, Hernández-Almeida I, Hoem F, Reilly B, Hemming S, Bailey I, Martos YM, Gutjahr M, Percuoco V, Allen C, Brachfeld S, Cardillo FG, Du Z, Fauth G, Fogwill C, Garcia M, Glüder A, Guitard M, Hwang J-H, Iizuka M, Kenlee B, O’Connell S, Pérez LF, Ronge TA, Seki O, Tauxe L, Tripathi S, Zheng X (2022) Ancient marine sediment DNA reveals diatom transition in Antarctica. Nat Commun 13(1):5787. https://doi.org/10.1038/s41467-022-33494-4
Barouillet C, Vasselon V, Keck F, Millet L, Etienne D, Galop D, Rius D, Domaizon I (2022) Paleoreconstructions of ciliate communities reveal long-term ecological changes in temperate lakes. Sci Rep 12(1):7899. https://doi.org/10.1038/s41598-022-12041-7
Barouillet C, Monchamp M-E, Bertilsson S, Brasell K, Domaizon I, Epp LS, Ibrahim A, Mejbel H, Nwosu EC, Pearman JK, Picard M, Thomson-Laing G, Tsugeki N, Von Eggers J, Gregory-Eaves I, Pick FR, Wood SA, Capo E (2023) Investigating the effects of anthropogenic stressors on lake biota using sedimentary DNA. Freshw Biol. https://doi.org/10.1111/fwb.14027
Beisser D, Graupner N, Bock C, Wodniok S, Grossmann L, Vos M, Sures B, Rahmann S, Boenigk J (2017) Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes. PeerJ 5:e2832. https://doi.org/10.7717/peerj.2832
Bik HM, Porazinska DL, Creer S, Caporaso JG, Knight R, Thomas WK (2012) Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol Evol 27(4):233–243. https://doi.org/10.1016/j.tree.2011.11.010
Bjorbækmo MFM, Evenstad A, Røsæg LL, Krabberød AK, Logares R (2020) The planktonic protist interactome: where do we stand after a century of research? ISME J 14(2):544–559. https://doi.org/10.1038/s41396-019-0542-5
Boenigk J, Arndt H (2002) Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Van Leeuwenhoek 81(1):465–480. https://doi.org/10.1023/A:1020509305868
Boere AC, Abbas B, Rijpstra WIC, Versteegh GJM, Volkman JK, Sinninghe Damsté JS, Coolen MJL (2009) Late-Holocene succession of dinoflagellates in an Antarctic fjord using a multi-proxy approach: paleoenvironmental genomics, lipid biomarkers and palynomorphs. Geobiology 7(3):265–281. https://doi.org/10.1111/j.1472-4669.2009.00202.x
Boere AC, Rijpstra WIC, De Lange GJ, Sinninghe Damsté JS, Coolen MJL (2011a) Preservation potential of ancient plankton DNA in Pleistocene marine sediments. Geobiology 9(5):377–393. https://doi.org/10.1111/j.1472-4669.2011.00290.x
Boere AC, Sinninghe Damsté JS, Rijpstra WIC, Volkman JK, Coolen MJL (2011b) Source-specific variability in post-depositional DNA preservation with potential implications for DNA based paleoecological records. Org Geochem 42(10):1216–1225. https://doi.org/10.1016/j.orggeochem.2011.08.005
Brasell KA, Pochon X, Howarth J, Pearman JK, Zaiko A, Thompson L, Vandergoes MJ, Simon KS, Wood SA (2022) Shifts in DNA yield and biological community composition in stored sediment: implications for paleogenomic studies. Metabarcoding Metagenom 6:e78128. https://doi.org/10.3897/mbmg.6.78128
Bråte J, Logares R, Berney C, Ree DK, Klaveness D, Jakobsen KS, Shalchian-Tabrizi K (2010) Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J 4(9):1144–1153. https://doi.org/10.1038/ismej.2010.39
Burki F (2014) The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6(5):a016147. https://doi.org/10.1101/cshperspect.a016147
Burki F, Roger AJ, Brown MW, Simpson AGB (2020) The new tree of eukaryotes. Trends Ecol Evol 35(1):43–55. https://doi.org/10.1016/j.tree.2019.08.008
Burki F, Sandin MM, Jamy M (2021) Diversity and ecology of protists revealed by metabarcoding. Curr Biol 31(19):R1267–R1280. https://doi.org/10.1016/j.cub.2021.07.066
Capo E, Debroas D, Arnaud F, Domaizon I (2015) Is planktonic diversity well recorded in sedimentary DNA? Toward the reconstruction of past protistan diversity. Microb Ecol 70(4):865–875. https://doi.org/10.1007/s00248-015-0627-2
Capo E, Debroas D, Arnaud F, Guillemot T, Bichet V, Millet L, Gauthier E, Massa C, Develle A-L, Pignol C, Lejzerowicz F, Domaizon I (2016) Long-term dynamics in microbial eukaryotes communities: a palaeolimnological view based on sedimentary DNA. Mol Ecol 25(23):5925–5943. https://doi.org/10.1111/mec.13893
Capo E, Debroas D, Arnaud F, Perga M-E, Chardon C, Domaizon I (2017a) Tracking a century of changes in microbial eukaryotic diversity in lakes driven by nutrient enrichment and climate warming. Environ Microbiol 19(7):2873–2892. https://doi.org/10.1111/1462-2920.13815
Capo E, Domaizon I, Maier D, Debroas D, Bigler C (2017b) To what extent is the DNA of microbial eukaryotes modified during burying into lake sediments? A repeat-coring approach on annually laminated sediments. J Paleolimnol 58(4):479–495. https://doi.org/10.1007/s10933-017-0005-9
Capo E, Rydberg J, Tolu J, Domaizon I, Debroas D, Bindler R, Bigler C (2019) How does environmental inter-annual variability shape aquatic microbial communities? A 40-year annual record of sedimentary DNA from a boreal Lake (Nylandssjön, Sweden). Front Ecol Evol 7
Capo E, Giguet-Covex C, Rouillard A, Nota K, Heintzman PD, Vuillemin A, Ariztegui D, Arnaud F, Belle S, Bertilsson S, Bigler C, Bindler R, Brown AG, Clarke CL, Crump SE, Debroas D, Englund G, Ficetola GF, Garner RE, Gauthier J, Gregory-Eaves I, Heinecke L, Herzschuh U, Ibrahim A, Kisand V, Kjær KH, Lammers Y, Littlefair J, Messager E, Monchamp M-E, Olajos F, Orsi W, Pedersen MW, Rijal DP, Rydberg J, Spanbauer T, Stoof-Leichsenring KR, Taberlet P, Talas L, Thomas C, Walsh DA, Wang Y, Willerslev E, van Woerkom A, Zimmermann HH, Coolen MJL, Epp LS, Domaizon IG, Alsos I, Parducci L (2021a) Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: overview and recommendations. Quaternary 4(1):6. https://doi.org/10.3390/quat4010006
Capo E, Ninnes S, Domaizon I, Bertilsson S, Bigler C, Wang X-R, Bindler R, Rydberg J (2021b) Landscape setting drives the microbial eukaryotic community structure in four Swedish Mountain lakes over the Holocene. Microorganisms 9(2):355. https://doi.org/10.3390/microorganisms9020355
Capo E, Monchamp M-E, Coolen MJL, Domaizon I, Armbrecht L, Bertilsson S (2022) Environmental paleomicrobiology: using DNA preserved in aquatic sediments to its full potential. Environ Microbiol 24(5):2201–2209. https://doi.org/10.1111/1462-2920.15913
Caron DA, Hu SK (2019) Are we overestimating protistan diversity in nature? Trends Microbiol 27(3):197–205. https://doi.org/10.1016/j.tim.2018.10.009
Caron DA, Hutchins DA (2013) The effects of changing climate on microzooplankton grazing and community structure: drivers, predictions and knowledge gaps. J Plankton Res 35(2):235–252. https://doi.org/10.1093/plankt/fbs091
Chambouvet A, Berney C, Romac S, Audic S, Maguire F, De Vargas C, Richards TA (2014) Diverse molecular signatures for ribosomally ‘active’ Perkinsea in marine sediments. BMC Microbiol 14(1):110. https://doi.org/10.1186/1471-2180-14-110
Ciobanu M-C, Burgaud G, Dufresne A, Breuker A, Rédou V, Ben Maamar S, Gaboyer F, Vandenabeele-Trambouze O, Lipp JS, Schippers A, Vandenkoornhuyse P, Barbier G, Jebbar M, Godfroy A, Alain K (2014) Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J 8(7):1370–1380. https://doi.org/10.1038/ismej.2013.250
Coolen MJL, Shtereva G (2009) Vertical distribution of metabolically active eukaryotes in the water column and sediments of the Black Sea. FEMS Microbiol Ecol 70(3):525–539. https://doi.org/10.1111/j.1574-6941.2009.00756.x
Coolen MJL, Muyzer G, Rijpstra WIC, Schouten S, Volkman JK, Sinninghe Damsté JS (2004) Combined DNA and lipid analyses of sediments reveal changes in Holocene haptophyte and diatom populations in an Antarctic lake. Earth Planet Sci Lett 223:225–239. https://doi.org/10.1016/j.epsl.2004.04.014
Coolen MJL, Orsi WD, Balkema C, Quince C, Harris K, Sylva SP, Filipova-Marinova M, Giosan L (2013) Evolution of the plankton paleome in the Black Sea from the Deglacial to Anthropocene. Proc Natl Acad Sci 110(21):8609–8614. https://doi.org/10.1073/pnas.1219283110
Corinaldesi C, Danovaro R, Dell’Anno A (2005) Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl Environ Microbiol 71(1):46–50. https://doi.org/10.1128/AEM.71.1.46-50.2005
Debroas D, Domaizon I, Humbert J-F, Jardillier L, Lepère C, Oudart A, Taïb N (2017) Overview of freshwater microbial eukaryotes diversity: a first analysis of publicly available metabarcoding data. FEMS Microbiol Ecol 93(4):fix023. https://doi.org/10.1093/femsec/fix023
Delebecq G, Schmidt S, Ehrhold A, Latimier M, Siano R (2020) Revival of ancient marine dinoflagellates using molecular biostimulation. J Phycol 56(4):1077–1089. https://doi.org/10.1111/jpy.13010
Dell’Anno A, Danovaro R (2005) Extracellular DNA plays a key role in Deep-Sea ecosystem functioning. Science 309(5744):2179–2179. https://doi.org/10.1126/science.1117475
DeLong JP, Van Etten JL, Al-Ameeli Z, Agarkova IV, Dunigan DD (2023) The consumption of viruses returns energy to food chains. Proc Natl Acad Sci 120(1):e2215000120. https://doi.org/10.1073/pnas.2215000120
Domaizon I, Winegardner A, Capo E, Gauthier J, Gregory-Eaves I (2017) DNA-based methods in paleolimnology: new opportunities for investigating long-term dynamics of lacustrine biodiversity. J Paleolimnol 58(1):1–21. https://doi.org/10.1007/s10933-017-9958-y
Dulias K, Stoof-Leichsenring KR, Pestryakova LA, Herzschuh U (2017) Sedimentary DNA versus morphology in the analysis of diatom-environment relationships. J Paleolimnol 57(1):51–66. https://doi.org/10.1007/s10933-016-9926-y
Edgcomb VP, Beaudoin D, Gast R, Biddle JF, Teske A (2011) Marine subsurface eukaryotes: the fungal majority. Environ Microbiol 13(1):172–183. https://doi.org/10.1111/j.1462-2920.2010.02318.x
Ellegaard M, Clokie MRJ, Czypionka T, Frisch D, Godhe A, Kremp A, Letarov A, McGenity TJ, Ribeiro S, John Anderson N (2020) Dead or alive: sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun Biol 3(1):1–11. https://doi.org/10.1038/s42003-020-0899-z
Filker S, Sommaruga R, Vila I, Stoeck T (2016) Microbial eukaryote plankton communities of high-mountain lakes from three continents exhibit strong biogeographic patterns. Mol Ecol 25(10):2286–2301. https://doi.org/10.1111/mec.13633
Finlay BJ, Fenchel T (1999) Divergent perspectives on protist species richness. Protist 150(3):229–233. https://doi.org/10.1016/S1434-4610(99)70025-8
Finlay BJ, Fenchel T (2004) Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 155(2):237–244. https://doi.org/10.1078/143446104774199619
Foissner W (1999) Protist diversity: estimates of the near-imponderable. Protist 150(3):363–368. https://doi.org/10.1016/S1434-4610(99)70037-4
Fonseca BM, Câmara PEAS, Ogaki MB, Pinto OHB, Lirio JM, Coria SH, Vieira R, Carvalho-Silva M, Amorim ET, Convey P, Rosa LH (2022) Green algae (Viridiplantae) in sediments from three lakes on Vega Island, Antarctica, assessed using DNA metabarcoding. Mol Biol Rep 49(1):179–188. https://doi.org/10.1007/s11033-021-06857-1
Forster D, Dunthorn M, Mahé F, Dolan JR, Audic S, Bass D, Bittner L, Boutte C, Christen R, Claverie J-M, Decelle J, Edvardsen B, Egge E, Eikrem W, Gobet A, Kooistra WHCF, Logares R, Massana R, Montresor M, Not F, Ogata H, Pawlowski J, Pernice MC, Romac S, Shalchian-Tabrizi K, Simon N, Richards TA, Santini S, Sarno D, Siano R, Vaulot D, Wincker P, Zingone A, de Vargas C, Stoeck T (2016) Benthic protists: the under-charted majority. FEMS Microbiol Ecol 92(8):fiw120. https://doi.org/10.1093/femsec/fiw120
Garner RE, Gregory-Eaves I, Walsh DA (2020) Sediment metagenomes as time capsules of lake microbiomes. mSphere 5(6):e00512-20. https://doi.org/10.1128/mSphere.00512-20
Garner RE, Kraemer SA, Onana VE, Huot Y, Gregory-Eaves I, Walsh DA (2022) Protist diversity and metabolic strategy in freshwater lakes are shaped by trophic state and watershed land use on a continental scale. mSystems 7(4):e00316-22. https://doi.org/10.1128/msystems.00316-22
Gauthier J, Walsh D, Selbie DT, Bourgeois A, Griffiths K, Domaizon I, Gregory-Eaves I (2021) Evaluating the congruence between DNA-based and morphological taxonomic approaches in water and sediment trap samples: analyses of a 36-month time series from a temperate monomictic lake. Limnol Oceanogr 66(8):3020–3039. https://doi.org/10.1002/lno.11856
Gauthier J, Walsh D, Selbie DT, Domaizon I, Gregory-Eaves I (2022) Sedimentary DNA of a human-impacted lake in Western Canada (Cultus Lake) reveals changes in micro-eukaryotic diversity over the past 200 years. Environmental DNA 4(5):1106–1119. https://doi.org/10.1002/edn3.310
Giner CR, Forn I, Romac S, Logares R, de Vargas C, Massana R (2016) Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol 82(15):4757–4766. https://doi.org/10.1128/AEM.00560-16
Grattepanche JD, Walker LM, Ott BM, Paim Pinto DL, Delwiche, CF, Lane CE, Katz LA (2018) Microbial Diversity in the Eukaryotic SAR Clade: Illuminating the Darkness Between Morphology and Molecular Data. Biological Sciences: Faculty Publications, Smith College, Northampton, MA. https://scholarworks.smith.edu/bio_facpubs/91
Grossmann L, Jensen M, Heider D, Jost S, Glücksman E, Hartikainen H, Mahamdallie SS, Gardner M, Hoffmann D, Bass D, Boenigk J (2016) Protistan community analysis: key findings of a large-scale molecular sampling. ISME J 10(9):2269–2279. https://doi.org/10.1038/ismej.2016.10
Hadziavdic K, Lekang K, Lanzen A, Jonassen I, Thompson EM, Troedsson C (2014) Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One 9(2):e87624. https://doi.org/10.1371/journal.pone.0087624
Härnström K, Ellegaard M, Andersen TJ, Godhe A (2011) Hundred years of genetic structure in a sediment revived diatom population. Proc Natl Acad Sci 108(10):4252–4257. https://doi.org/10.1073/pnas.1013528108
Hou W, Dong H, Li G, Yang J, Coolen MJL, Liu X, Wang S, Jiang H, Wu X, Xiao H, Lian B, Wan Y (2014) Identification of photosynthetic plankton communities using sedimentary ancient DNA and their response to late-Holocene climate change on the Tibetan plateau. Sci Rep 4(1):6648. https://doi.org/10.1038/srep06648
Huang S, Herzschuh U, Pestryakova LA, Zimmermann HH, Davydova P, Biskaborn BK, Shevtsova I, Stoof-Leichsenring KR (2020) Genetic and morphologic determination of diatom community composition in surface sediments from glacial and thermokarst lakes in the Siberian Arctic. J Paleolimnol 64(3):225–242. https://doi.org/10.1007/s10933-020-00133-1
Huo S, Zhang H, Monchamp M-E, Wang R, Weng N, Zhang J, Zhang H, Wu F (2022a) Century-long homogenization of algal communities is accelerated by nutrient enrichment and climate warming in lakes and reservoirs of the north temperate zone. Environ Sci Technol 56(6):3780–3790. https://doi.org/10.1021/acs.est.1c06958
Huo S, Zhang H, Wang J, Chen J, Wu F (2022b) Temperature and precipitation dominates millennium changes of eukaryotic algal communities in Lake Yamzhog Yumco, southern Tibetan Plateau. Sci Total Environ 829:154636. https://doi.org/10.1016/j.scitotenv.2022.154636
Ibrahim A, Capo E, Wessels M, Martin I, Meyer A, Schleheck D, Epp LS (2021) Anthropogenic impact on the historical phytoplankton community of lake constance reconstructed by multimarker analysis of sediment-core environmental DNA. Mol Ecol 30(13):3040–3056. https://doi.org/10.1111/mec.15696
Jones RI (2000) Mixotrophy in planktonic protists: an overview. Freshw Biol 45(2):219–226. https://doi.org/10.1046/j.1365-2427.2000.00672.x
Kammerlander B, Breiner H-W, Filker S, Sommaruga R, Sonntag B, Stoeck T (2015) High diversity of protistan plankton communities in remote high mountain lakes in the European Alps and the Himalayan mountains. FEMS Microbiol Ecol 91(4):fiv010. https://doi.org/10.1093/femsec/fiv010
Kanbar HJ, Olajos F, Englund G, Holmboe M (2020) Geochemical identification of potential DNA-hotspots and DNA-infrared fingerprints in lake sediments. Appl Geochem 122:104728. https://doi.org/10.1016/j.apgeochem.2020.104728
Keck F, Millet L, Debroas D, Etienne D, Galop D, Rius D, Domaizon I (2020) Assessing the response of micro-eukaryotic diversity to the great acceleration using lake sedimentary DNA. Nat Commun 11(1):3831. https://doi.org/10.1038/s41467-020-17682-8
Kirkpatrick J, Walsh E, D’Hondt S (2016) Fossil DNA persistence and decay in marine sediment over hundred-thousand-year to million-year time scales. Graduate School of Oceanography Faculty Publications https://doi.org/10.1130/G37933.1
Kisand V, Talas L, Kisand A, Stivrins N, Reitalu T, Alliksaar T, Vassiljev J, Liiv M, Heinsalu A, Seppä H, Veski S (2018) From microbial eukaryotes to metazoan vertebrates: wide spectrum paleo-diversity in sedimentary ancient DNA over the last ~14,500 years. Geobiology 16(6):628–639. https://doi.org/10.1111/gbi.12307
Kjær KH, Winther Pedersen M, De Sanctis B, De Cahsan B, Korneliussen TS, Michelsen CS, Sand KK, Jelavić S, Ruter AH, Schmidt AMA, Kjeldsen KK, Tesakov AS, Snowball I, Gosse JC, Alsos IG, Wang Y, Dockter C, Rasmussen M, Jørgensen ME, Skadhauge B, Prohaska A, Kristensen JÅ, Bjerager M, Allentoft ME, Coissac E, Rouillard A, Simakova A, Fernandez-Guerra A, Bowler C, Macias-Fauria M, Vinner L, Welch JJ, Hidy AJ, Sikora M, Collins MJ, Durbin R, Larsen NK, Willerslev E (2022) A 2-million-year-old ecosystem in Greenland uncovered by environmental DNA. Nature 612(7939):283–291. https://doi.org/10.1038/s41586-022-05453-y
Klouch KZ, Schmidt S, Andrieux-Loyer F, Le Gac M, Hervio-Heath D, Qui-Minet ZN, Quéré J, Bigeard E, Guillou L, Siano R (2016) Historical records from dated sediment cores reveal the multidecadal dynamic of the toxic dinoflagellate Alexandrium minutum in the Bay of Brest (France). FEMS Microbiol Ecol 92(7):fiw101. https://doi.org/10.1093/femsec/fiw101
Krantzberg G (1985) The influence of bioturbation on physical, chemical and biological parameters in aquatic environments: a review. Environ Pollut Ser A, Ecol Biol 39(2):99–122. https://doi.org/10.1016/0143-1471(85)90009-1
Lammers Y, Heintzman PD, Alsos IG (2021) Environmental palaeogenomic reconstruction of an ice age algal population. Commun Biol 4(1):1–11. https://doi.org/10.1038/s42003-021-01710-4
Lefranc M, Thénot A, Lepère C, Debroas D (2005) Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl Environ Microbiol 71(10):5935–5942. https://doi.org/10.1128/AEM.71.10.5935-5942.2005
Lepère C, Domaizon I, Debroas D (2008) Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl Environ Microbiol 74(10):2940–2949. https://doi.org/10.1128/AEM.01156-07
Li G, Dong H, Hou W, Wang S, Jiang H, Yang J, Wu G (2016) Temporal succession of ancient phytoplankton community in Qinghai lake and implication for paleo-environmental change. Sci Rep 6(1):19769. https://doi.org/10.1038/srep19769
Li M, Bastos Gomes G, Zhao W, Hu G, Huang K, Yoshinaga T, Clark TG, Li W, Zou H, Wu S, Wang G (2023) Cultivation of fish ciliate parasites: progress and prospects. Rev Aquac 15(1):142–162. https://doi.org/10.1111/raq.12708
Likens GE (ed) (2010) Plankton of inland waters: a derivative of encyclopedia of inland waters. Academic Press/Elsevier, San Diego
Lischke B, Weithoff G, Wickham SA, Attermeyer K, Grossart HP, Scharnweber K, HIlt S, Gaedke U (2016) Large biomass of small feeders: ciliates may dominate herbivory in eutrophic lakes. J Plankton Res 38(1):2–15. https://doi.org/10.1093/plankt/fbv102
Lynn DH, Doerder FP, Gillis PL, Prosser RS (2018) Tetrahymena glochidiophila n. sp., a new species of Tetrahymena (Ciliophora) that causes mortality to glochidia larvae of freshwater mussels (Bivalvia). Dis Aquat Org 127(2):125–136. https://doi.org/10.3354/dao03188
Mangot J-F, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, Debroas D (2013) Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ Microbiol 15(6):1745–1758. https://doi.org/10.1111/1462-2920.12065
Martin JL, Santi I, Pitta P, John U, Gypens N (2022) Towards quantitative metabarcoding of eukaryotic plankton: an approach to improve 18S rRNA gene copy number bias. Metabarcoding Metagenom 6:e85794. https://doi.org/10.3897/mbmg.6.85794
Marzetz V, Spijkerman E, Striebel M, Wacker A (2020) Phytoplankton community responses to interactions between light intensity, light variations, and phosphorus supply. Front Environ Sci 8
McCarthy FMG, Mertens KN, Ellegaard M, Sherman K, Pospelova V, Ribeiro S, Blasco S, Vercauteren D (2011) Resting cysts of freshwater dinoflagellates in southeastern Georgian Bay (Lake Huron) as proxies of cultural eutrophication. Rev Palaeobot Palynol 166(1):46–62. https://doi.org/10.1016/j.revpalbo.2011.04.008
Millette NC, Gast RJ, Luo JY, Moeller HV, Stamieszkin K, Andersen KH, Brownlee EF, Cohen NR, Duhamel S, Dutkiewicz S, Glibert PM, Johnson MD, Leles SG, Maloney AE, Mcmanus GB, Poulton N, Princiotta SD, Sanders RW, Wilken S (2023) Mixoplankton and mixotrophy: future research priorities. J Plankton Res:fbad020. https://doi.org/10.1093/plankt/fbad020
Mitra A, Flynn KJ, Burkholder JM, Berge T, Calbet A, Raven JA, Granéli E, Glibert PM, Hansen PJ, Stoecker DK, Thingstad F, Tillmann U, Våge S, Wilken S, Zubkov MV (2014) The role of mixotrophic protists in the biological carbon pump. Biogeosciences 11(4):995–1005. https://doi.org/10.5194/bg-11-995-2014
Mitra A, Flynn KJ, Tillmann U, Raven JA, Caron D, Stoecker DK, Not F, Hansen PJ, Hallegraeff G, Sanders R, Wilken S, McManus G, Johnson M, Pitta P, Våge S, Berge T, Calbet A, Thingstad F, Jeong HJ, Burkholder J, Glibert PM, Granéli E, Lundgren V (2016) Defining planktonic Protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167(2):106–120. https://doi.org/10.1016/j.protis.2016.01.003
Miyazono A, Nagai S, Kudo I, Tanizawa K (2012) Viability of Alexandrium tamarense cysts in the sediment of Funka Bay, Hokkaido, Japan: over a hundred year survival times for cysts. Harmful Algae 16:81–88. https://doi.org/10.1016/j.hal.2012.02.001
Moguel B, Pérez L, Alcaraz LD, Blaz J, Caballero M, Muñoz-Velasco I, Becerra A, Laclette JP, Ortega-Guerrero B, Romero-Oliva CS, Herrera-Estrella L, Lozano-García S (2021) Holocene life and microbiome profiling in ancient tropical Lake Chalco. Mexico Sci Rep 11(1):13848. https://doi.org/10.1038/s41598-021-92981-8
More KD, Orsi WD, Galy V, Giosan L, He L, Grice K, Coolen MJL (2018) A 43 kyr record of protist communities and their response to oxygen minimum zone variability in the Northeastern Arabian Sea. Earth Planet Sci Lett 496:248–256. https://doi.org/10.1016/j.epsl.2018.05.045
Nanney DL, Park C, Preparata R, Simon EM (1998) Comparison of sequence differences in a variable 23S rRNA domain among sets of cryptic species of ciliated protozoa. J Eukaryot Microbiol 45(1):91–100. https://doi.org/10.1111/j.1550-7408.1998.tb05075.x
Nolte V, Pandey RV, Jost S, Medinger R, Ottenwälder B, Boenigk J, Schlötterer C (2010) Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol Ecol 19(14):2908–2915. https://doi.org/10.1111/j.1365-294X.2010.04669.x
Oikonomou A, Filker S, Breiner H-W, Stoeck T (2015) Protistan diversity in a permanently stratified meromictic Lake (lake Alatsee, SW Germany). Environ Microbiol 17(6):2144–2157. https://doi.org/10.1111/1462-2920.12666
Orlando L, Allaby R, Skoglund P, Der Sarkissian C, Stockhammer PW, Ávila-Arcos MC, Fu Q, Krause J, Willerslev E, Stone AC, Warinner C (2021) Ancient DNA analysis. Nat Rev Methods Primers 1(1):1–26. https://doi.org/10.1038/s43586-020-00011-0
Orsi W, Biddle JF, Edgcomb V (2013) Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8(2):e56335. https://doi.org/10.1371/journal.pone.0056335
Pachiadaki MG, Rédou V, Beaudoin DJ, Burgaud G, Edgcomb VP (2016) Fungal and prokaryotic activities in the marine subsurface biosphere at Peru margin and Canterbury Basin inferred from RNA-based analyses and microscopy. Front Microbiol 7
Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M, Fiore-Donno AM, Gile GH, Holzmann M, Jahn R, Jirků M, Keeling PJ, Kostka M, Kudryavtsev A, Lara E, Lukeš J, Mann DG, Mitchell EAD, Nitsche F, Romeralo M, Saunders GW, Simpson AGB, Smirnov AV, Spouge JL, Stern RF, Stoeck T, Zimmermann J, Schindel D, Vargas C de (2012) CBOL Protist Working Group: Barcoding Eukaryotic Richness beyond the Animal, Plant, and Fungal Kingdoms. PLOS Biology 10(11):e1001419. https://doi.org/10.1371/journal.pbio.1001419
Pawlowski J, Kelly-Quinn M, Altermatt F, Apothéloz-Perret-Gentil L, Beja P, Boggero A, Borja A, Bouchez A, Cordier T, Domaizon I, Feio MJ, Filipe AF, Fornaroli R, Graf W, Herder J, van der Hoorn B, Iwan Jones J, Sagova-Mareckova M, Moritz C, Barquín J, Piggott JJ, Pinna M, Rimet F, Rinkevich B, Sousa-Santos C, Specchia V, Trobajo R, Vasselon V, Vitecek S, Zimmerman J, Weigand A, Leese F, Kahlert M (2018) The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci Total Environ 637–638:1295–1310. https://doi.org/10.1016/j.scitotenv.2018.05.002
Pernthaler J, Posch T (2009) Microbial food webs. In: Likens GE (ed) Encyclopedia of inland waters. Academic, Oxford, pp 244–251
Pick FR, Caron DA (1987) Picoplankton and Nanoplankton biomass in Lake Ontario: relative contribution of phototrophic and heterotrophic communities. Can J Fish Aquat Sci 44:2164–2172. https://doi.org/10.1139/f87-265
Posch T, Köster O, Salcher MM, Pernthaler J (2012) Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nature Clim Change 2(11):809–813. https://doi.org/10.1038/nclimate1581
Prosser RS, Lynn DH, Salerno J, Bennett J, Gillis PL (2018) The facultatively parasitic ciliated protozoan, Tetrahymena glochidiophila (Lynn, 2018), causes a reduction in viability of freshwater mussel glochidia. J Invertebr Pathol 157:25–31. https://doi.org/10.1016/j.jip.2018.07.012
Randlett M-È, Coolen MJL, Stockhecke M, Pickarski N, Litt T, Balkema C, Kwiecien O, Tomonaga Y, Wehrli B, Schubert CJ (2014) Alkenone distribution in Lake Van sediment over the last 270 ka: influence of temperature and haptophyte species composition. Quat Sci Rev 104:53–62. https://doi.org/10.1016/j.quascirev.2014.07.009
Sagova-Mareckova M, Boenigk J, Bouchez A, Cermakova K, Chonova T, Cordier T, Eisendle U, Elersek T, Fazi S, Fleituch T, Frühe L, Gajdosova M, Graupner N, Haegerbaeumer A, Kelly A-M, Kopecky J, Leese F, Nõges P, Orlic S, Panksep K, Pawlowski J, Petrusek A, Piggott JJ, Rusch JC, Salis R, Schenk J, Simek K, Stovicek A, Strand DA, Vasquez MI, Vrålstad T, Zlatkovic S, Zupancic M, Stoeck T (2021) Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring. Water Res 191:116767. https://doi.org/10.1016/j.watres.2020.116767
Sanders RW (2009) Protists. In: Likens GE (ed) Encyclopedia of inland waters. Academic, Oxford, pp 252–260
Sandgren CD, Smol JP, Kristiansen J (eds) (1995) Chrysophyte algae: ecology, phylogeny and development. Cambridge University Press, Cambridge
Sanyal A, Larsson J, van Wirdum F, Andrén T, Moros M, Lönn M, Andrén E (2022) Not dead yet: diatom resting spores can survive in nature for several millennia. Am J Bot 109(1):67–82. https://doi.org/10.1002/ajb2.1780
Seeber PA, von Hippel B, Kauserud H, Löber U, Stoof-Leichsenring KR, Herzschuh U, Epp LS (2022) Evaluation of lake sedimentary ancient DNA metabarcoding to assess fungal biodiversity in Arctic paleoecosystems. Environmental DNA 4(5):1150–1163. https://doi.org/10.1002/edn3.315
Sherr EB, Sherr BF (2002) Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81(1):293–308. https://doi.org/10.1023/A:1020591307260
Siano R, Lassudrie M, Cuzin P, Briant N, Loizeau V, Schmidt S, Ehrhold A, Mertens KN, Lambert C, Quintric L, Noël C, Latimier M, Quéré J, Durand P, Penaud A (2021) Sediment archives reveal irreversible shifts in plankton communities after world war II and agricultural pollution. Curr Biol 31(12):2682–2689.e7. https://doi.org/10.1016/j.cub.2021.03.079
Singer D, Seppey CVW, Lentendu G, Dunthorn M, Bass D, Belbahri L, Blandenier Q, Debroas D, de Groot GA, de Vargas C, Domaizon I, Duckert C, Izaguirre I, Koenig I, Mataloni G, Schiaffino MR, Mitchell EAD, Geisen S, Lara E (2021) Protist taxonomic and functional diversity in soil, freshwater and marine ecosystems. Environ Int 146:106262. https://doi.org/10.1016/j.envint.2020.106262
Smol JP (1985) The ratio of diatom frustules to chrysophycean statospores: a useful paleolimnological index. Hydrobiologia 123(3):199–208. https://doi.org/10.1007/BF00034378
Smol JP, Birks HJB, Last WM, Bradley RS, Alverson K (eds) (2001) Tracking environmental change using Lake sediments: terrestrial, algal, and siliceous indicators. Springer, Dordrecht
Stoof-Leichsenring KR, Epp LS, Trauth MH, Tiedemann R (2012) Hidden diversity in diatoms of Kenyan Lake Naivasha: a genetic approach detects temporal variation. Mol Ecol 21(8):1918–1930. https://doi.org/10.1111/j.1365-294X.2011.05412.x
Stoof-Leichsenring KR, Bernhardt N, Pestryakova LA, Epp LS, Herzschuh U, Tiedemann R (2014) A combined paleolimnological/genetic analysis of diatoms reveals divergent evolutionary lineages of Staurosira and Staurosirella (Bacillariophyta) in Siberian lake sediments along a latitudinal transect. J Paleolimnol 52(1):77–93. https://doi.org/10.1007/s10933-014-9779-1
Stoof-Leichsenring KR, Herzschuh U, Pestryakova LA, Klemm J, Epp LS, Tiedemann R (2015) Genetic data from algae sedimentary DNA reflect the influence of environment over geography. Sci Rep 5(1):12924. https://doi.org/10.1038/srep12924
Stoof-Leichsenring KR, Liu S, Jia W, Li K, Pestryakova LA, Mischke S, Cao X, Liu X, Ni J, Neuhaus S, Herzschuh U (2020a) Plant diversity in sedimentary DNA obtained from high-latitude (Siberia) and high-elevation lakes (China). Biodivers Data J 8:e57089. https://doi.org/10.3897/BDJ.8.e57089
Stoof-Leichsenring KR, Pestryakova LA, Epp LS, Herzschuh U (2020b) Phylogenetic diversity and environment form assembly rules for Arctic diatom genera – a study on recent and ancient sedimentary DNA. J Biogeogr 47(5):1166–1179. https://doi.org/10.1111/jbi.13786
Suthers I, Rissik D, Richardson A (eds) (2019) Plankton: a guide to their ecology and monitoring for water quality. CSIRO publishing
Talas L, Stivrins N, Veski S, Tedersoo L, Kisand V (2021) Sedimentary ancient DNA (sedaDNA) reveals fungal diversity and environmental drivers of community changes throughout the Holocene in the present boreal Lake Lielais Svētiņu (eastern Latvia). Microorganisms 9(4):719. https://doi.org/10.3390/microorganisms9040719
Tõnno I, Talas L, Freiberg R, Kisand A, Belle S, Stivrins N, Alliksaar T, Heinsalu A, Veski S, Kisand V (2021) Environmental drivers and abrupt changes of phytoplankton community in temperate lake Lielais Svētiņu, eastern Latvia, over the last post-glacial period from 14.5 kyr. Quat Sci Rev 263:107006. https://doi.org/10.1016/j.quascirev.2021.107006
Triadó-Margarit X, Casamayor EO (2012) Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain). Environ Microbiol 14(9):2445–2456. https://doi.org/10.1111/j.1462-2920.2012.02797.x
Triadó-Margarit X, Casamayor EO (2015) High protists diversity in the plankton of sulfurous lakes and lagoons examined by 18s rRNA gene sequence analyses. Environ Microbiol Rep 7(6):908–917. https://doi.org/10.1111/1758-2229.12324
Vasselon V, Rimet F, Domaizon I, Monnier O, Reyjol Y, Bouchez A (2019) Assessing pollution of aquatic environments with diatoms’ DNA metabarcoding: experience and developments from France water framework directive networks. Metabarcoding Metagenom 3:e39646. https://doi.org/10.3897/mbmg.3.39646
Weisbrod B, Wood SA, Steiner K, Whyte-Wilding R, Puddick J, Laroche O, Dietrich DR (2020) Is a central sediment sample sufficient? Exploring spatial and temporal microbial diversity in a small lake. Toxins 12(9):580. https://doi.org/10.3390/toxins12090580
Weisse T, Montagnes DJS (2022) Ecology of planktonic ciliates in a changing world: concepts, methods, and challenges. J Eukaryot Microbiol 69(5):e12879. https://doi.org/10.1111/jeu.12879
Wetzel RG (2001) Limnology: Lake and river ecosystems. Elsevier
Wurzbacher C, Fuchs A, Attermeyer K, Frindte K, Grossart H-P, Hupfer M, Casper P, Monaghan MT (2017) Shifts among Eukaryota, bacteria, and archaea define the vertical organization of a lake sediment. Microbiome 5(1):41. https://doi.org/10.1186/s40168-017-0255-9
Xiong W, Jousset A, Li R, Delgado-Baquerizo M, Bahram M, Logares R, Wilden B, de Groot GA, Amacker N, Kowalchuk GA, Shen Q, Geisen S (2021) A global overview of the trophic structure within microbiomes across ecosystems. Environ Int 151:106438. https://doi.org/10.1016/j.envint.2021.106438
Zhang H, Huo S, Cao X, Ma C, Zhang J, Wu F (2021) Homogenization of reservoir eukaryotic algal and cyanobacterial communities is accelerated by dam construction and eutrophication. J Hydrol 603:126842. https://doi.org/10.1016/j.jhydrol.2021.126842
Zimmermann HH, Stoof-Leichsenring KR, Dinkel V, Harms L, Schulte L, Hütt M-T, Nürnberg D, Tiedemann R, Herzschuh U (2023) Marine ecosystem shifts with deglacial sea-ice loss inferred from ancient DNA shotgun sequencing. Nat Commun 14(1):1650. https://doi.org/10.1038/s41467-023-36845-x
Zinger L, Bonin A, Alsos IG, Bálint M, Bik H, Boyer F, Chariton AA, Creer S, Coissac E, Deagle BE, De Barba M, Dickie IA, Dumbrell AJ, Ficetola GF, Fierer N, Fumagalli L, Gilbert MTP, Jarman S, Jumpponen A, Kauserud H, Orlando L, Pansu J, Pawlowski J, Tedersoo L, Thomsen PF, Willerslev E, Taberlet P (2019) DNA metabarcoding – need for robust experimental designs to draw sound ecological conclusions. Mol Ecol 28(8):1857–1862. https://doi.org/10.1111/mec.15060
Acknowledgements
We acknowledge financial support from the Pole Research & Development on Lacustrine Ecosystems (ECLA) of the French Biodiversity Agency (OFB) and the Severo Ochoa Excellence Program postdoctoral fellowship awarded in 2021 to Eric Capo (CEX2019-000928-S). We are very grateful to Kuldeep More, Thomas Weisse and Frances Pick for their critical reading of our chapter.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Barouillet, C., Domaizon, I., Capo, E. (2023). Protist DNA from Lake Sediments. In: Capo, E., Barouillet, C., Smol, J.P. (eds) Tracking Environmental Change Using Lake Sediments. Developments in Paleoenvironmental Research, vol 21. Springer, Cham. https://doi.org/10.1007/978-3-031-43799-1_6
Download citation
DOI: https://doi.org/10.1007/978-3-031-43799-1_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43798-4
Online ISBN: 978-3-031-43799-1
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)